Around 1620 the Flemish chemist Jan Baptist van Helmont, often considered the father of pneumatic chemistry (the chemistry of gases), wrote the following:
"If you press a piece of underwear soiled with sweat together with some wheat in an open mouth jar, after about 21 days the odor changes and the ferment coming out of the underwear and penetrating through the husks of the wheat, changes the wheat into mice.''
This reflected the commonly held belief at that time, even among many scientists, of spontaneous generation. Life was assumed to arise spontaneously and continuously: mice from wheat, maggots from meat, frogs from mud, and so on.More than two centuries later, in 1862, Louis Pasteur won a price from the French Academy of Sciences for definitively putting to rest this idea of spontaneous generation. Pasteur earned his prize with a simple but clever experiment. He took several flasks and filled them with a sterilized broth. Then he closed the necks of the flasks and let them stand for several weeks. Checking back regularly, he observed that nothing happened. But as soon as he broke the neck of some of his flasks, exposing the broth to the open air, life (in the form of micro-organisms) would appear within several days. Pasteur thus concluded that all life comes from other life.

Just a few years earlier, in 1859, Charles Darwin had published his book On the Origin of Species. One of the main ideas underlying his theory of evolution by natural selection is that of common descent: any (arbitrary) group of currently living species will, if you go far enough back in time, have a common ancestor. For example, humans and chimps have a common ancestor that lived around 6-7 million years ago, and all currently living bird species have their common ancestry within a group of dinosaurs known as theropoda, 145-200 million years ago. Similarly, all life on earth must have come from one (or just a few) common ancestor(s).So, if all life comes from life, going all the way back to a "last universal common ancestor'' (LUCA), then where did this common ancestor, one of the very first living organisms, come from? The origin of life had become a genuine scientific problem.
A Selfish RNA World
The main paradigm in current origin of life research is that of an RNA world. RNA is a similar kind of molecule as DNA, but mostly exists in single-stranded form. So, instead of forming a rather inert double helix structure like DNA, RNA molecules fold into a complicated 3-dimensional structure, which allows them to be chemically active. Moreover, some RNA molecules can act as catalysts for chemical reactions between other RNA molecules.A catalyst is a molecule that speeds up the rate at which a reaction happens, without being used up in that reaction. Catalysis is ubiquitous in living systems, and life probably could not exist without it. Catalysts are essential in determining and regulating the functionality of the chemical reaction networks that support life.Initially, the origin of life problem revolved around the chicken-and-egg question of "which came first: DNA or proteins?" DNA molecules store genetic information, which is translated into proteins that act as catalysts. But some of these protein catalysts, in turn, are necessary to replicate and translate DNA. Hence the catch-22.

However, it seems that RNA can fulfill both roles simultaneously. It can store genetic information because of its similarity to DNA, and it can act as a catalyst because of its chemically active structure, like proteins. Furthermore, next to DNA and proteins, RNA also plays an important role in the molecular machinery underlying living systems. For example, the ribosome, the main DNA-protein translational apparatus, consists primarily of RNA. All of this gave rise to the idea of an RNA world: life starting with a few self-replicating RNA molecules that were responsible for both the replication and expression of genetic information.Such an RNA world would be a "selfish" world, where each RNA molecule is responsible for its own replication, and where different RNA molecules would be competing for resources (basic building blocks, such as individual nucleotides). However, despite the attractiveness and simplicity of the idea, so far nobody has been able to show that RNA can indeed catalyze its own template-directed replication.
Cooperative Molecular Networks
What has been shown experimentally, though, is that some RNA molecules can catalyze the formation of other RNA molecules from shorter RNA fragments. Moreover, there are experimentally constructed sets of RNA molecules that mutually catalyze each other's formation. In other words, rather than having each RNA molecule replicate itself, they all help each other's formation from their basic building blocks, in a "cooperative" molecular network.The first example of such a mutually catalytic network was constructed in the lab of Günter von Kiedrowski, and consists of a cross-catalytic set of short nucleotide sequences. The basic building blocks are the trimers A=CCG and B=CGG, which form each other's base-pair complement when read in opposite directions. The hexamers AA and BB now serve as templates to which the complementary trimers can attach by forming C-G base-pair bonds. For example, two B trimers can attach to an AA template, allowing these trimers to ligate (chemically join) into a fully formed BB hexamer. After strand separation, the original AA template is regained, plus a new BB template. In a similar way, such a BB template can facilitate the ligation of another AA template from two A trimers. This chemical reaction network is shown schematically in the figure below.

These ligation reactions could, in principle, also happen spontaneously. They would happen at very low rates, though, as the trimers would have to line up in exactly the right way, by chance, for the ligation reaction to proceed. However, the hexamers (templates) facilitate this reaction by having the trimers attach to them and thus aligning them in exactly the right way. After the ligation reaction has happened and the two hexamers have separated, the original template is regained. In other words, the two hexamers truly catalyze each other's formation from their basic building blocks.Later on, a similar cross-catalytic set of two much longer RNA molecules (more than 70 nucleotides long) was constructed experimentally in the lab of Gerald Joyce. Moreover, these RNA sequences were subjected to an artificial form of evolution, significantly increasing their catalytic efficiency. More recently, several experimental systems with up to 16 RNA molecules (of around 200 nucleotides) that mutually catalyze each other's formation from shorter fragments were created in the lab of Niles Lehman. However, such experimental systems are not restricted to RNA molecules alone. A similar set of nine mutually catalytic peptides (short proteins) was created and studied in detail by Gonen Ashkenasy and colleagues.
Autocatalytic Sets
What these experimental systems of mutually catalytic molecules have in common, is that they are all instances of an autocatalytic set. An autocatalytic setis defined as a chemical reaction network, i.e., a set of reactions and the molecules involved in them, such that:
- each reaction in the set is catalyzed by at least one of the molecules from the set itself, and
- each molecule in the set can be built up from a basic food source through a sequence of reactions from the set itself.
The food source consists of the basic building blocks, such as the RNA or peptide fragments in the experimental examples of autocatalytic sets described above, or the molecules that happened to be present on the early earth in a purely prebiotic setting. In other words, the food source consists of those elements that can be assumed to be available in the environment, and that can act as reactants or catalysts, but which do not necessarily have to be produced by the reaction network itself.Note that this concept of an autocatalytic set captures two essential properties of living systems: all chemical reactions are facilitated and regulated by catalysts generated within the network itself (i.e., it is catalytically closed, condition 1 in the definition above), and it is self-sustaining from resources available in the environment (condition 2).The figure below shows a simple example of an autocatalytic set formed by a reaction network where the molecules (black dots) are represented by bit strings (i.e., strings of zeros and ones, or "binary polymers"), the food source consists of the monomers and dimers (i.e., the bit strings of lengths one and two), and the longer molecules (polymers) can be built up through ligation reactions (white boxes) between two shorter bit strings. Solid black arrows indicate reactants going into and products coming out of a (ligation) reaction, and dashed gray arrows indicate which molecules catalyze which reactions. Given the definition above, it is easy to verify that this reaction network satisfies its two conditions, and thus indeed forms an autocatalytic set.

This concept of autocatalytic sets was originally introduced by Stuart Kauffman, in descriptive form already back in 1971, and more formally in 1986. However, more detailed studies of autocatalytic sets were mostly done over the past decade or so.
Properties of Autocatalytic Sets
What these detailed studies have shown, is that autocatalytic sets have a high probability of existing in computational models of chemical reaction networks, also for chemically plausible levels of catalysis. For example, in the binary polymer model described above, where catalysis is assigned randomly, each molecule only needs to catalyze between one and two reactions, on average, to already have a high probability for autocatalytic sets to exist. These results do not change much when more realistic ways of assigning catalysts are used, such as when a (potential) catalyst must match a certain number of bits around the ligation site, similar to the C-G base-pair bonding on which the catalysis in von Kiedrowski's original experimental system is based.Furthermore, it turns out that autocatalytic sets often consist of a hierarchy of smaller and smaller autocatalytic subsets. In other words, a given autocatalytic set often contains several smaller subsets that themselves also form autocatalytic sets. For example, in the figure above, which shows an autocatalytic set of five reactions, there exist two smaller subsets (one of two reactions and one of three reactions) that by themselves also form autocatalytic sets. Another example is presented in the figure below, which shows a larger autocatalytic set (also from the binary polymer model), where the various autocatalytic subsets are indicated by the differently colored shapes.

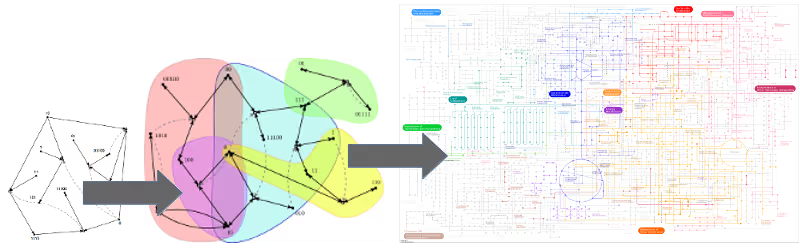
This property could enable the existence of different types of "protocells". Imagine two compartments formed by lipid membranes, which have been shown to form, grow, and divide spontaneously under appropriate circumstances. Now assume that the same "binary polymer chemistry" can take place in both of these compartments, but in one of them only the red autocatalytic subset is currently present, and in the other one only the blue autocatalytic subset. This would thus form two different types of protocells, which might then compete with each other for food resources (in this case the monomers and dimers), and perhaps even give rise to some simple evolutionary dynamics. Computational studies have shown that autocatalytic sets are indeed able to evolve and become more complex over time, exactly because of this existence of multiple autocatalytic subsets, which can come into existence at various times and in different combinations due to occasional spontaneous reactions.Of course these results are mostly based on simple computational models of chemical reaction networks, such as the binary polymer model. However, the formal autocatalytic sets framework has also been used to study some of the existing experimental networks in more detail. For example, many of the experimental observations in Lehman's 16-member RNA autocatalytic set were reproduced by the formal framework. In addition, several new insights were gained from this formal analysis, which would have been very difficult, or even impossible, to get from the experiments alone. Similar formal analyses are currently underway with Ashkenasy's peptide autocatalytic set.Furthermore, it was shown that the metabolic network of a well-known bacterium, E. coli, forms a large autocatalytic set. This, of course, supports the original claim that autocatalytic sets capture essential properties of living systems, in particular their catalytic closure and self-sustainability. In conclusion, autocatalytic sets are not just abstract mathematical constructs, but they do exist in real chemical and biological reaction networks, and can be studied formally within those systems.
The Origin of Life as a Cooperative Effort
Given that autocatalytic sets are highly likely to exist (also for chemically plausible levels of catalysis), that they are able to evolve and become more complex, and that they exist in real chemical and biological networks, an alternative scenario emerges for a possible origin of life. Perhaps life started with the spontaneous formation of one or more autocatalytic sets, which then gradually diversified and evolved into more and more complex chemical networks, eventually leading to truly autonomous, free living cells.Of course in modern-day organisms the chemical reactions that happen inside them (i.e., organic reactions) are catalyzed by highly specific and efficient enzymes. However, there is good evidence that at least several important organic reactions can also be catalyzed by inorganic elements such as various minerals and metals. Of course these inorganic elements are much less efficient than modern-day enzymes, but at the origin of life any catalysis, even if relatively inefficient, would have provided an advantage over no catalysis at all. Besides, most of these inorganic elements would have been able to catalyze many different reactions, as opposed to highly specific enzymes which can generally only catalyze just one or a few reactions (although with much higher efficiency).Furthermore, many of these inorganic elements are still used as cofactors in modern-day enzymes. Enzymes usually consist of long proteins that are folded up in a complicated way. Their 3-dimensional structure allows them to bind to specific substrates, holding them in the right place so they can undergo a chemical reaction. However, many enzymes also hold yet another element, often a small molecule like a mineral or metal, which acts as the actual catalyst for that reaction. This additional element is called the enzyme's cofactor. The protein simply makes this cofactor more efficient and specific by binding only to a relatively small set of substrates, and holding them all in exactly the right place so they can react properly. It seems plausible that an enzyme's cofactor would have been the original catalyst for that reaction.Finally, there is increasing evidence that RNA molecules and peptides interacted very early on in the origin and evolution of life (rather than an RNA world eventually giving rise to peptides), and that even short peptides can already have significant catalytic capabilities. Given that life depends on a diversity of molecule types, which all interact with each other in complicated ways, it seems hard to imagine how it could have started with just a single type of molecule.
All of this provides and alternative, metabolism-focused view of a possible origin of life. Starting with the spontaneous emergence of small autocatalytic sets of various molecule types, in which some of the original catalysts would have been simple inorganic elements and perhaps some short RNAs and peptides, these reaction networks would then have evolved and become more complex, creating more efficient catalysts (small enzymes incorporating cofactors) that take over from the original, less efficient ones, and so on in an upward spiral of diversity, efficiency, and complexity. In other words, life arising as a cooperative effort among diverse molecule types in catalytically closed and self-sustaining chemical reaction networks. I believe there is grandeur in this view of (the origin of) life.